Building a global atlas of wildlife disease data
In 1922, Lewis Fry Richardson — a pioneer of the mathematics underlying meteorology — lamented that “perhaps some day in the dim future it will be possible to advance the computations faster than the weather advances and at a cost less than the saving to mankind due to the information gained. But that is a dream.” A century later, meteorology has entirely surpassed Richardson’s vision; the tools of modern science can perform once-impossible tasks, like identifying the signal of climate change in the weather on any day, anywhere in the world.
The core of this revolution was data synthesis: a global network of weather stations, and the billions of data points they collect every year, have allowed scientists to predict weather across settings ranging from the mundane (a 10-day forecast) to local emergencies (hurricane trajectories) and global crises (climate change impacts).
The meteorological revolution is often cited both as evidence of the benefits of massive data, and as an explanation for why, in the absence of that kind of data, outbreak prediction is so difficult. But the COVID-19 pandemic has created an unprecedented consensus that the world cannot wait any longer for the disease data revolution. At least a dozen efforts have been proposed to take the first steps on the human end (e.g., the World Health Organization’s Hub for Epidemic and Pandemic Intelligence, the U.S. CDC’s new Center for Forecasting and Outbreak Analytics, a Global Immunological Observatory), the wildlife end (the Global Virome Project), and the interface between them (the NIAID CREID Network and USAID’s PREDICT, STOP Spillover, and DEEP VZN programs).
This network of programs will generate and aggregate previously-unthinkable volumes of data, but none of these are primarily designed to address where that data completes its life cycle. Data on human and wildlife disease is a precious commodity, and the sharing of these data can be mired in issues around academic incentives, transparency, ethical disclosures, informed consent, access and benefit sharing, and biosecurity risks. Only recently have these conversations pointed towards the possibility of standardizing global data sharing on human disease (e.g., the global.health project). But on the wildlife end - where most emerging diseases get started, and where hundreds of thousands more pathogens are lurking mostly undetected - no such system has been proposed.
Last year, the Verena team started building our open data ecosystem with The Global Virome in One Network (VIRION), an open synthetic database of vertebrate-virus species interactions. Starting in 2022, we’re tackling several new data projects, and today, we wanted to announce the biggest one in development: the Pathogen Harmonized Occurrence and Surveillance (PHAROS) database, a global open repository of animal disease detection data.
Our goal is to build a global atlas of wildlife disease, and we need your help.
Every pathogen, every animal, every record
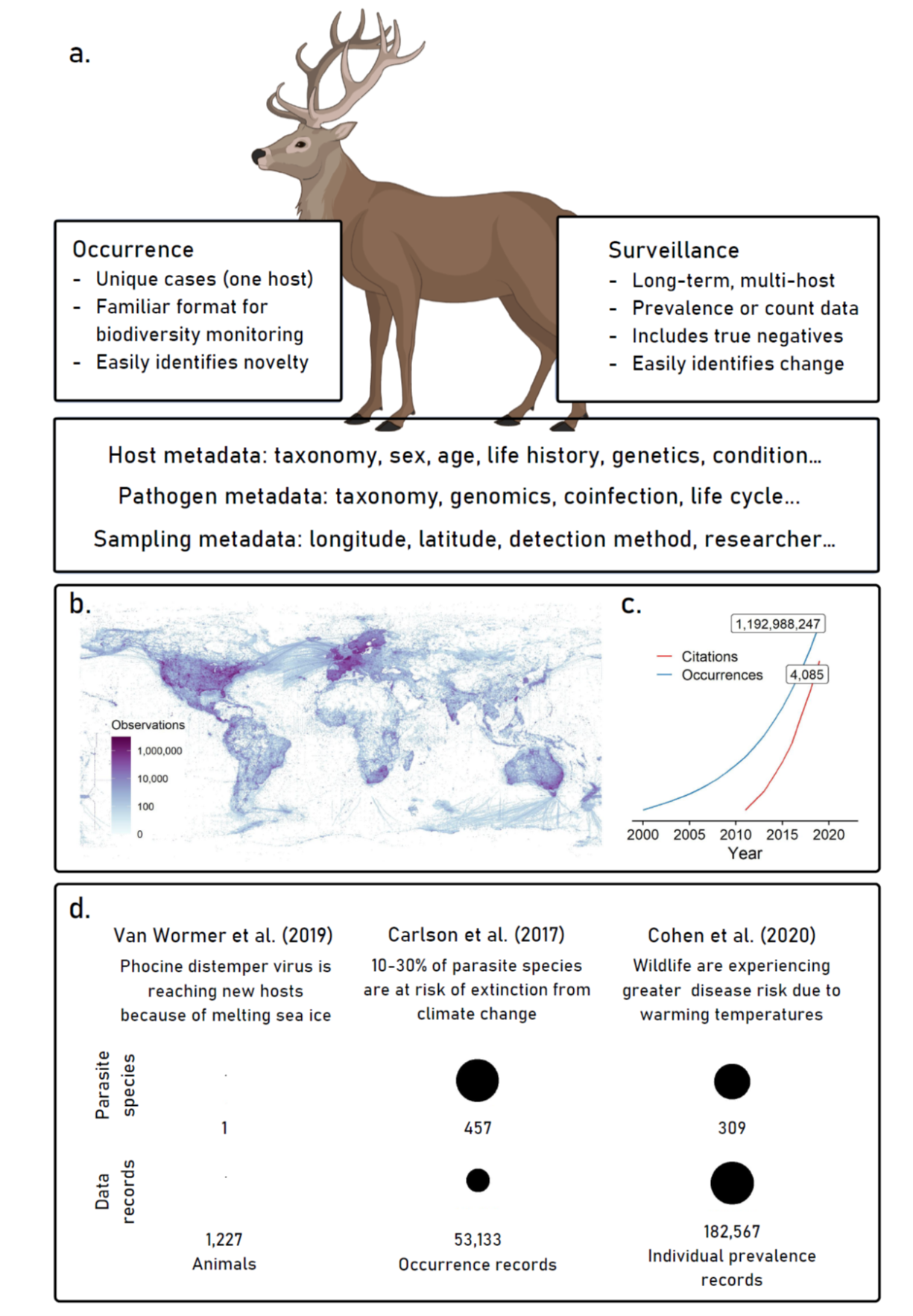
Millions of records of wildlife diseases are published every year in scientific journals and grey literature, unstandardized, unconsolidated, and inaccessible for reuse—a network of weather stations each broadcasting a different combination of sensors in their own bulletin, with a mix of formatting, languages, and detail. Researchers need somewhere to deposit their data, ideally one that can capture two basic reporting formats: occurrence (point data describing the observation of a parasite or pathogen) and surveillance (more detailed reporting of pathogen prevalence, incidence, or intensity). The former is already captured for some parasites in biodiversity repositories like the Global Biodiversity Informatics Facility (GBIF; www.gbif.org), particularly thanks to data shared by parasite collections around the world, but nearly every existing format is insufficient for the latter. Some efforts have standardized these data from the literature, such as the Global Mammal Parasite Database, but they lack the key feature that sparked the biodiversity data revolution: open user contributions, and community norms that encourage them as part of the standard scientific process of publication.
In service of this goal, we’re building a global collaborative effort around the goal of a One Health data synthesis revolution. To support that effort, we’ve been developing a flexible, open-access, community-maintained data hub where researchers can report any results from wildlife samples—from one record of a tick on an unexpected host, to a multi-decade study of bat virus seroprevalence. The primary function of PHAROS is consolidation and standardization, not data generation: researchers already report prevalence values in publications, log occurrence data on GBIF or with museum accessions, and deposit pathogen genetic sequence data (GSD) on GenBank or specialized platforms (e.g. GISAID for influenza; www.gisaid.org). By developing a primary platform for reporting the sampling component of these data, and linking to other platforms for GSD, we propose that these data can be standardized for reuse across purposes and platforms. Our project can also benefit from bioinformatic structures that make existing databases tractable, by adopting and adapting architectures like the NCBI taxonomic backbone (which we use in VIRION!). By doing so, we can make sure PHAROS stays compatible with other open datasets, and make it easier to update these sources together in real-time.
A tremendous of data specific to parasitism and infection are collected that are not usually stored in standard biodiversity data formats (A). Programs like GBIF (gbif.org) have accelerated data collection and synthesis in ecology (B), leading to an unprecedented snapshot of global biodiversity (C); we propose a similar effort to synthesize data on wildlife disease. The growing availability of these data has already led to massive scientific advances, for example, in our understanding of global climate change impacts (D); a global monitoring system would transform the possibilities of this work.
As a first step, we’ve been consolidating data already in the literature through manual and text-mining approaches to curation. If you have data you want to deposit, reach out—we’d love to host it.
Next, we’ll start developing a broader collaborative platform to promote, govern, and formalize data sharing. Building a global weather system is not as simple as collecting weather data: a million records of wildlife disease might already be available as grey data from published studies, but to gather a billion data points, we’ll need to build a research community that benefits from and is invested in PHAROS. As Verena grows, we’re bringing in new collaborators to help shape plans in terms of credit and attribution for contributors, long-term financial sustainability, and the value returned to the global community. (There’s plenty out there that we can learn from already, like GBIF’s DOIs for data downloads.)
One billion records
Our hope is that jumpstarting this data revolution will give epidemiologists new raw materials to develop models of disease transmission, and help microbiologists resolve the underpinnings of cross-species evolutionary jumps. The impacts of global change would also be easier to detect; even groundbreaking studies of climate change impacts on wildlife health are limited today to a few thousand data points at a time, and are usually the product of years of both data collection and analysis. With millions of records, scientists might be able to pinpoint when parasites reach higher latitudes, viruses cross species barriers for the first time, or warmer temperatures spark fungal epidemics, on timescales of just weeks to months. The science of early warning signals remains similarly young, and limited by data availability, but with high-resolution spatiotemporal data on disease dynamics, scientists might be able to advance the mathematics of spatial early warning signals to finally develop real-time systems that alert vulnerable populations to zoonotic spillover risk. At long last, outbreak forecasting might be just as achievable as a 10-day weather forecast – at a coarser scale, no doubt, but still to a degree that supports outbreak prevention on the human side of the spillover interface.
Reaching that point requires a level of cooperation that extends beyond normal data synthesis. No one team of researchers could collate these data into existence; they can only be generated if the community adopts norms about open data sharing, and programs engaged in disease surveillance at scale make data deposition a priority. At present, it could seem like this normative shift would undermine the value of these programs, and the benefit they provide to researchers’ careers. However, biodiversity science offers a key lesson on this front: as the amount of data in GBIF has grown, passing the thresholds of millions and billions of records, the proportional value of each unique record has marginally (if at all) decreased, but the cumulative value of the synthesized data to the global community has grown exponentially. The incentives to share data now far outweigh the marginal gains from suppressing them, and the universal curation of a global biodiversity dataset allows researchers anywhere in the world to start a scientific research project from any computer. Without this, our understanding of processes like climate change, deforestation, and the sixth mass extinction would have been massively reduced; hundreds of citations and entire paragraphs of evidence in IPCC or IPBES reports would simply be non-existent. This process is similarly vital for a post-pandemic effort to understand infectious diseases, and a global atlas of wildlife disease could empower researchers from all backgrounds to delve into questions about how disease works, how animal health is changing in an increasingly-fragile world, and how humans should face the growing threat of emerging pathogens.
We’re ready to get started—and we hope you’ll join us.