After a long hiatus… Bat βCoV Watch 🦇
(For the first time, in its permanent home!)
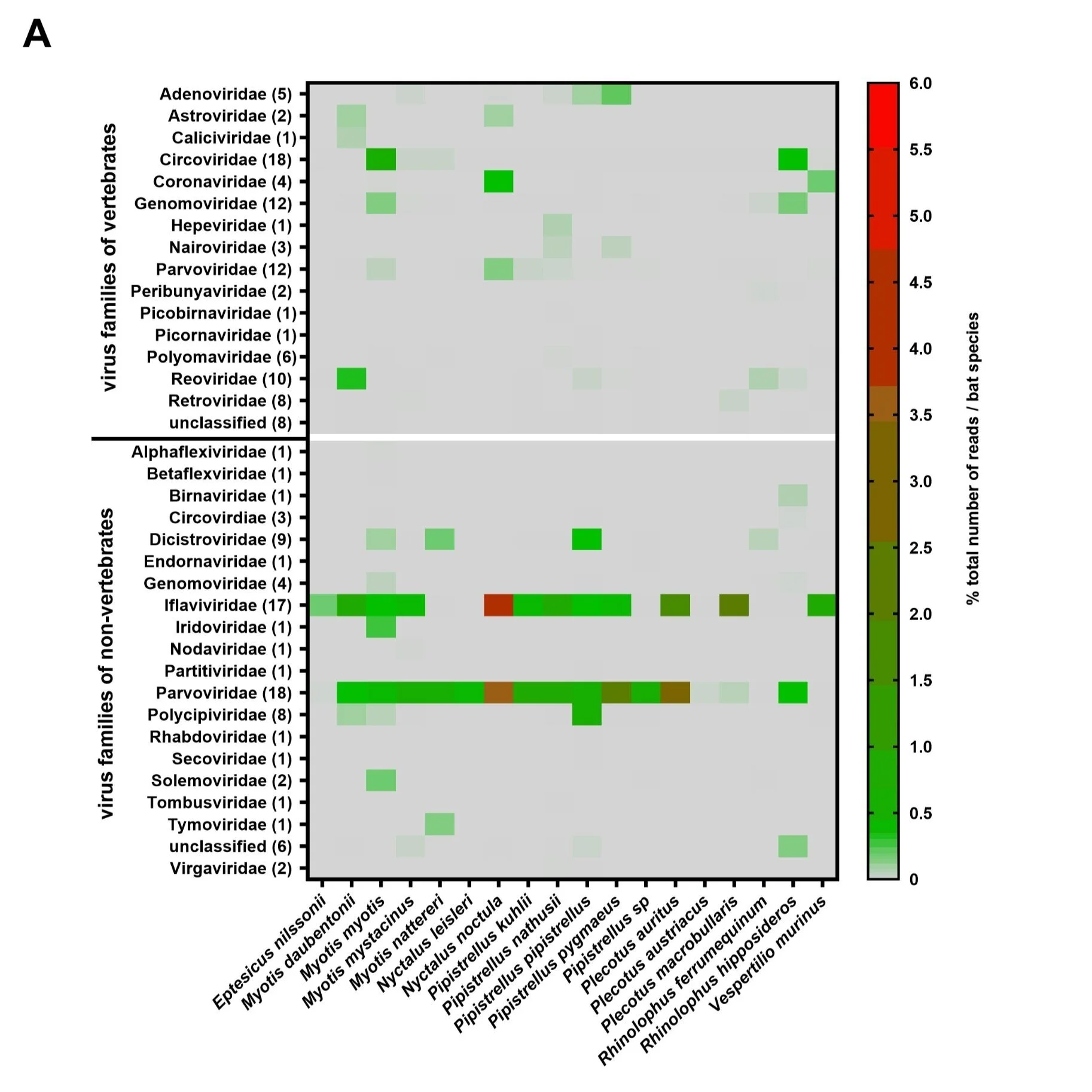
Image credit: Hardmeier et al. (2021, PLoS One)
Welcome back to Verena’s bat betacoronavirus watch, where we keep an eye out for the discovery of novel betacoronaviruses in bats. It’s pretty much what it says on the tin. We do this because:
We manually curate an updating database of the bats that are known hosts / reservoirs of betacoronaviruses, to support predictive modeling or other science.
We use that database to track the running accuracy of our predictive ensemble, so we know what works and what doesn’t (which means we can make better models).
While we’ve dispatched these updates over Twitter in the past, we’ve been planning to move this feature onto the blog for ages. Unfortunately, there’s been a small hitch: despite a number of interesting viral discoveries, we haven’t had any new hosts to report for ages.
That’s not for a lack of incredible science floating around - especially researchers digging into Rhinolophus samples. Here’s some watches-that-weren’t:
This study reported a new βCoV in Rhinolophus blythi, a species that - oddly enough - isn’t even in our database. That got our hopes up at first, when we thought it might be Blyth’s horseshoe bat (R. lepidus), but it turns out this probably refers to one of two other bats: R. subbadius, which would be a new data point for us, or more likely R. pusillus blythi, and pusillus is unfortunately a known host. (We dropped the authors an email just to check, but for now, not a new data point.)
SARS-like viruses were found in Rhinolophus species in Russia! Incredibly cool science that helps connect similar viruses found in Eastern Europe to those found in East Asia, but alas, no new hosts for us.
Some new bat viruses from Nigeria, including a SARS-like virus in Hipposideros ruber. and another betacoronavirus in Epomophorus gambianus, but again, no new hosts for us.
A number of viruses closely related to SARS-CoV-2 were found by this team in Yunnan province, China; excitingly, that includes RpYN06 from Rhinolophus pusillus, which is the closest relative to SARS-CoV-2 in most of the genome (outside of the spike protein) that’s been found in bats so far. Unfortunately, again, no new hosts for us.
The biggest payload: this incredible study in Hong Kong Med J. reported the results of samples from 9,866 bats (54 species) collected from 2004 to 2014. (This is a massive number of samples; for comparison, the PREDICT program reported the results of testing of 12,333 bat back in 2017.) There’s a number of interesting results, including viruses similar to both SARS-CoV and MERS-CoV, but somehow, in all of those samples, there’s not a single new βCoV-positive host for our database.
This has been, needless to say, a disappointing stretch for our team. But I’m excited to tell you the long, dark winter of βCoV watch is over today.
This uneventful period is brought to a close by Hardmeier et al., “Metagenomic analysis of fecal and tissue samples from 18 endemic bat species in Switzerland revealed a diverse virus composition including potentially zoonotic viruses.“ Amidst many other interesting results, this study reports the presence of a virus with 86% similarity to MERS-CoV in the parts of the genome they could assemble. That virus, BatCoV/V. murinus/Switzerland/2019, is our first record of a betacoronavirus in the Particoloured Bat (Vespertilio murinus), also endearingly known as the “rearmouse.”
V. murinus is one of the stranger cases of our model’s ensemble going a different direction than the components. Five models out of the eight predict this species as a positive (Trait-1, Trait-2, Trait-3, Network-1, and Network-3), but the ensemble’s strict thresholding leads it to miss the mark. Oh well!
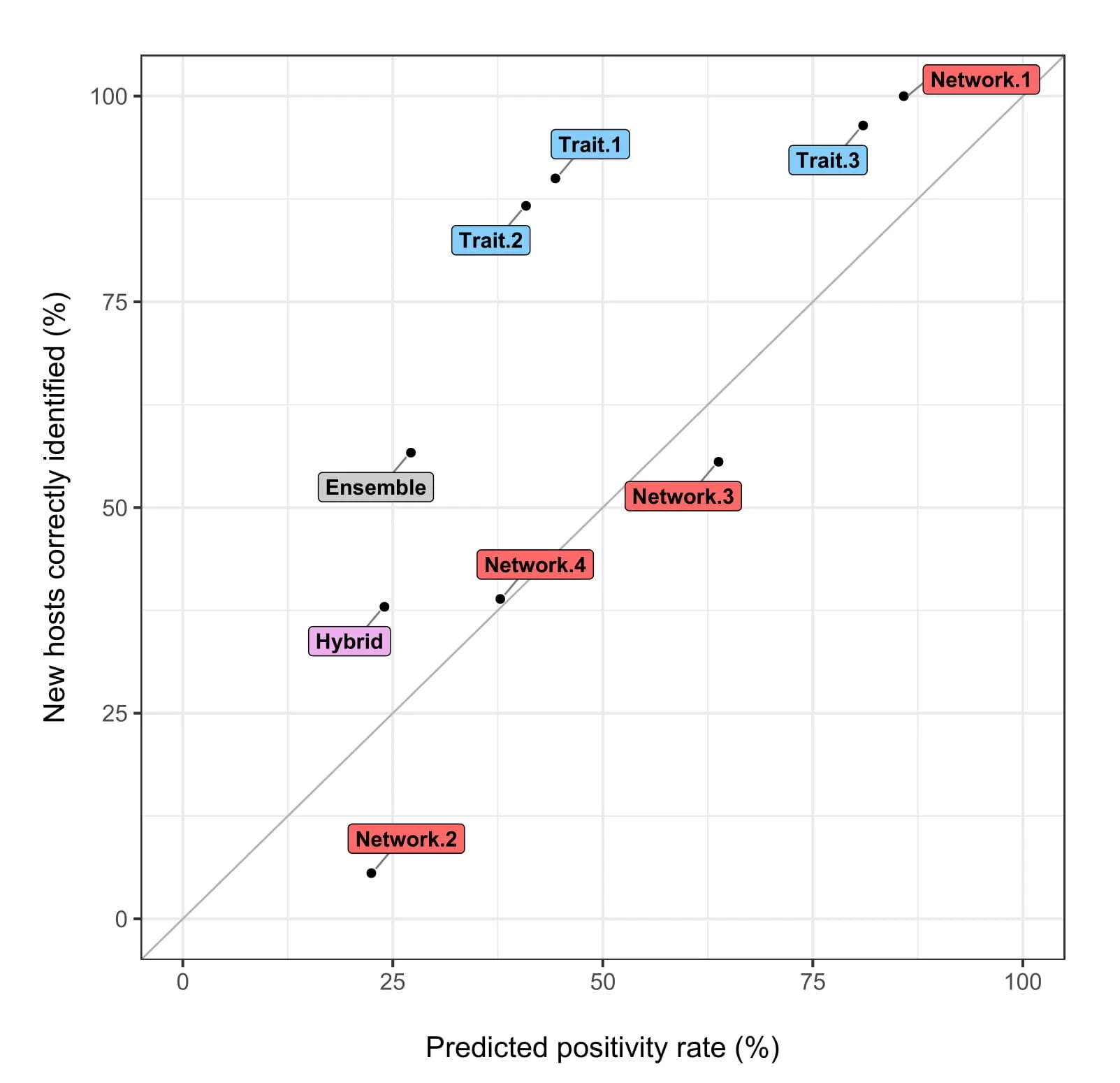
This new data point brings us to a landmark total of 30 new hosts since our original model runs. The ensemble correctly clocked 17 of those (57%); the leading model remains Network-1, with 100% accuracy (but only in-sample predictions), and Trait-3, with 96% accuracy (in- and out-of-sample). However, as we’ve learned, a model can achieve a high accuracy just by predicting that most bats are βCoV reservoirs, which is why we use our own custom diagnostic, the positivity-prediction curve:
In this diagnostic, the x axis shows the total % of bats predicted by the model to host these viruses, while the y axis shows the % correctly predicted from the 30 new hosts; a model performing at-random will fall on the grey line, while a good model will make as few suggestions as possible (low value on the x axis) with as high precision as possible (high value on the y axis). As it stands, Trait-1 and Trait-2 make the best predictions out of the narrowest suggested list, while the Network models mostly predict roughly-at-random. As we’ve learned over the last year, knowing about bat ecology helps us make more educated guesses about their role as viral reservoirs.
Thus concludes another Bat βCoV Watch. Thank you for your continued support of the Viral Emergence Research Initiative.
Dr. Colin Carlson
June 21, 2021